Outputs
Location
By default, fitting results are saved in the Output/ directory. You can specify a different directory using the --output (or -o) option:
chemex fit -o <path> [other options]
Multi-Step Fit
If the fitting method involves multiple steps, each step generates its own output subdirectory named after the fitting step.
Output/
├── STEP1/
└── STEP2/
Multi-Group Fit
In any fitting step, if the dataset can be divided into multiple groups (each with distinct fitting parameters), ChemEx fits each group separately. Results for each group are stored in numbered subfolders within the Groups/ folder, with a combined summary available in the All/ folder for convenience.
For example, if all global parameters (e.g., pB, kex) are fixed or if a residue-specific model (e.g., 2st_rs) is used, residue-specific fits are performed and organized as follows:
Output/
└── STEP2/
├── All/
└── Groups/
├── 10_11N/
├── 11_12N/
├── 12_13N/
Content
The output directory typically includes the following files and subdirectories:
Parameters/
Contains results of the fitting as three files: fitted.toml, fixed.toml, and constrained.toml, which list parameters that were fitted, fixed, and constrained, respectively.
Example Files
- fitted.toml
- fixed.toml
- constrained.toml
[GLOBAL]
KEX_AB = 3.81511e+02 # ±8.90870e+00
PB = 7.02971e-02 # ±1.14784e-03
[DW_AB]
15N = 2.00075e+00 # ±2.30817e-02
31N = 1.98968e+00 # ±1.90842e-02
Uncertainties (if calculated) are shown as comments preceded by "±", based on the covariance matrix from the Levenberg-Marquardt optimization.
[CS_A]
15N = 1.19849e+02 # (fixed)
31N = 1.26388e+02 # (fixed)
[GLOBAL]
KAB = 2.68192e+01 # ±3.06068e-01 ([KEX_AB] * [PB])
KBA = 3.54692e+02 # ±8.28245e+00 ([KEX_AB] * [PA])
Propagated uncertainties and applied constraints are provided in comments.
Plot/
Contains .pdf plots of the fitting results, along with the raw input and fitted data points. Example plots for CPMG and CEST experiments are shown below:
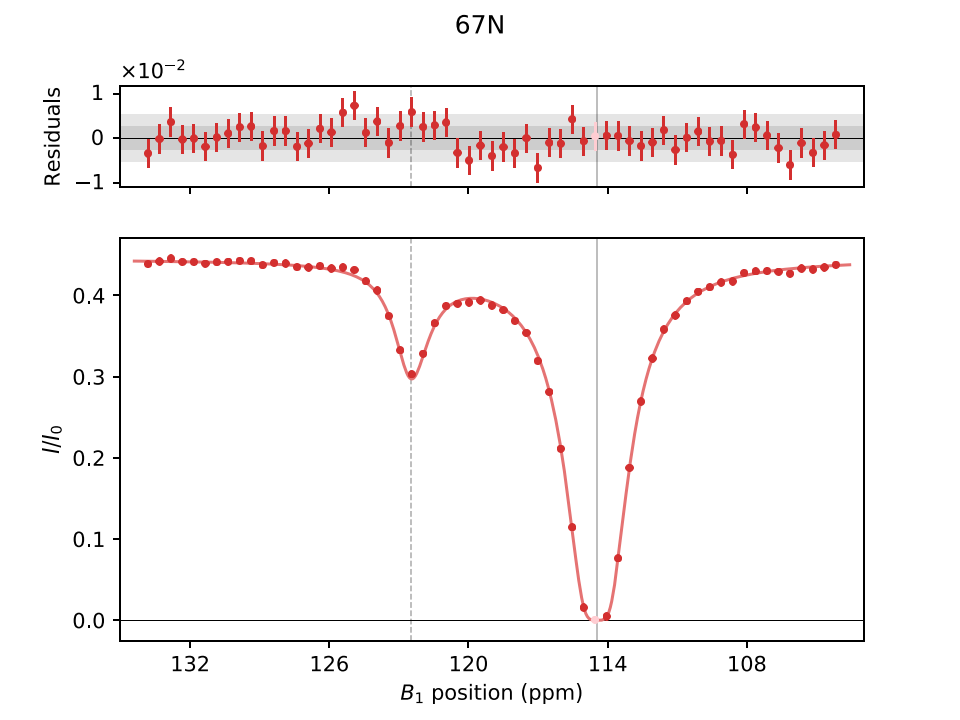
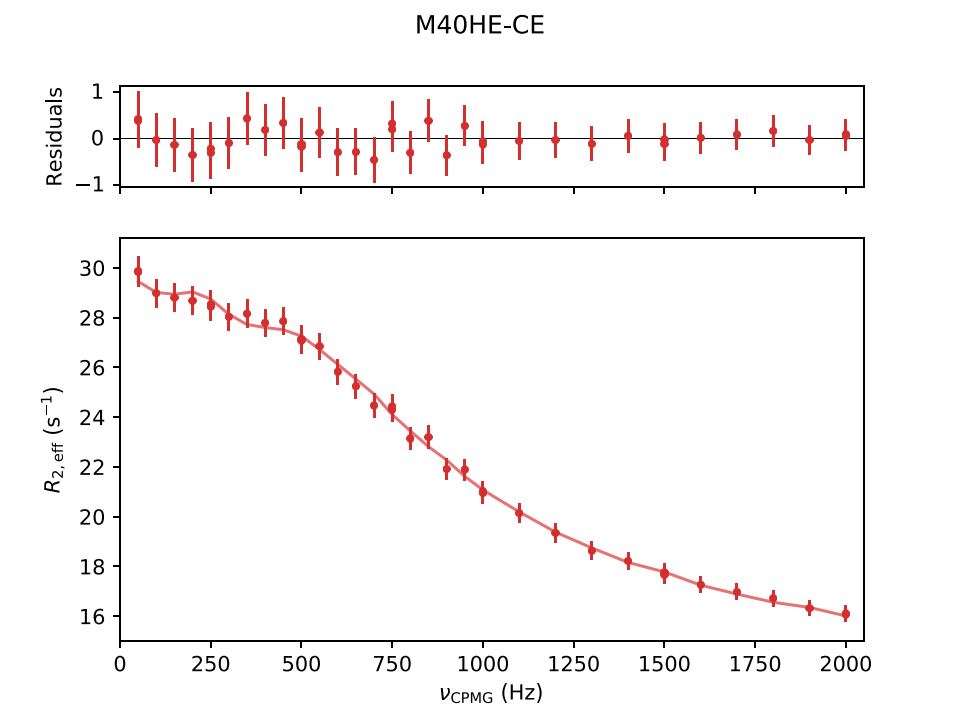
In (D-/cos-)CEST plots, solid and dashed vertical lines indicate ground and excited states, respectively, with lighter colors for filtered data points. "Folded" positions are marked with *.
Data/
Contains data values used in the fitting process along with back-calculated values for calculating χ2.
[15N]
# NCYC INTENSITY (EXP) ERROR (EXP) INTENSITY (CALC)
0 3.47059800e+04 1.77491406e+02 3.47055362e+04
statistics.toml
Contains goodness-of-fit statistics, such as χ2.
"number of data points" = 230
"number of variables" = 17
"chi-square" = 4.34824e+02
"reduced-chi-square" = 2.04143e+00
"Akaike Information Criterion (AIC)" = 1.80479e+02